Methods, Methods, Methods (My Research, Vol. 5)

On Tuesday, I can finally take the next big step in my project for my Bachelor thesis. Because of this, I wanted to show you some of the results I got so far, the long way you have to go before you can even do something that might lead to interesting results.
And while I was cropping pictures, adjusting colors and reducing image size, I realized one thing: Those of you, who don’t have a background in biology (aka: The majority of my readers) will have no idea what the hell I am even talking about.
Therefore, I decided to make a different post first.
This is said post. And it’s about methods. Methods I’ve been using, some of them are used on a daily basis in biology labs all around the world. I’ll try to make it as easy as possible (and thus not necessarily super-scientific), those who have already worked in a lab might want to skip this.
You have a piece of DNA. You want more pieces of DNA. Sadly, you don’t have any samples left to extract it from. That sucks. What do you do now?
Luckily, the replication of DNA is something all living things do all the time. There are certain enzymes that are specialized in making a lot of DNA out of a little bit of DNA. Scientists were able to isolate those enzymes and create a way to easily multiply DNA, without needing a living organism for that. The process is called PCR.@suesa
What happens during a PCR?
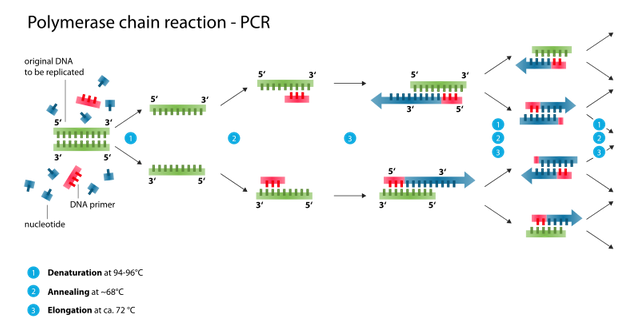 By Enzoklop (Own work) [CC BY-SA 3.0]
By Enzoklop (Own work) [CC BY-SA 3.0]
Thanks, Wikipedia. That was easy.
Okay, maybe not.
So, you have this double-stranded DNA (need a reminder what DNA is? Click here). You then heat it (= denaturation), so it splits into two strands. Tiny pieces of single-stranded DNA (= Primers) attach to these longer single strands (= annealing). Then, an enzyme called polymerase starts synthesizing a new strand of DNA, matching the one that already exists (= elongation). When that is done, you have two double strands instead of the initial one.
And then you repeat. Over and over and over again.
This gets you a lot of DNA, pretty fast. Neat, isn’t it?
The only method I’m using more frequently than the PCR is gel electrophoresis. It’s good for checking the size of a DNA fragment or sorting DNA fragments after size.
For that, you first need a gel. The gels usually consist of agarose (a sugar) and some kind of buffer. Both are mixed, heated, and then poured in a form to let cool. There’s also a small comb inside the form, which creates tiny “pockets” inside the gel. That’s where your samples are put into.
When the gel is hard (okay, it’s usually kind of wobbly), it’s put in a chamber which is filled with the same buffer the gel was made with. Then, you put your samples into the pockets. And then … you turn on the electricity!
DNA is charged slightly negative, which will make it move towards positive charges. This is used to make the DNA “wander” through the gel, as at the upper end, there’s a negative charge and at the lower end a positive one. Bit by bit, the DNA wanders through the gel. Bigger fragments are a lot slower than smaller fragments, which allows a separation by size.
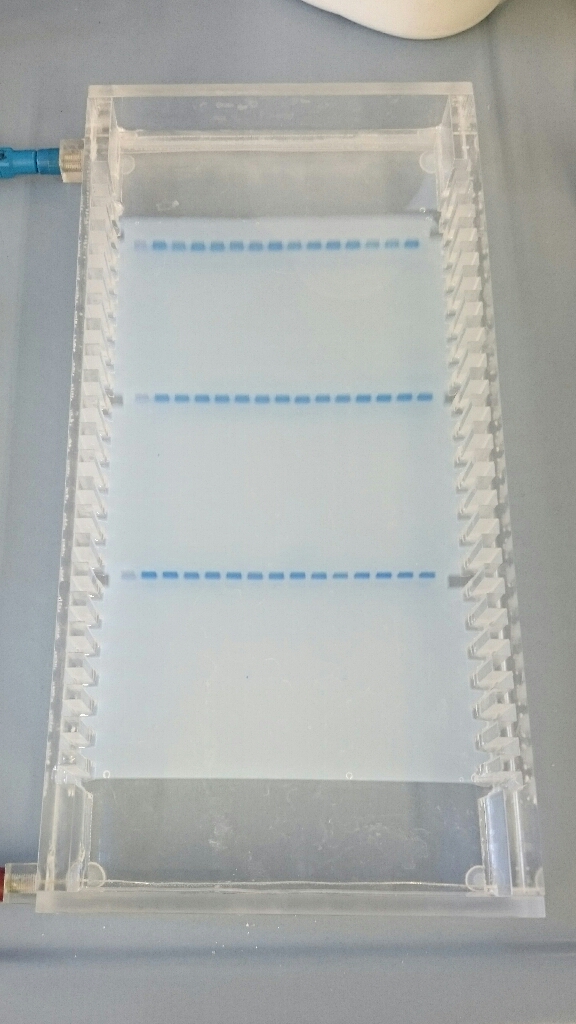
You just need to keep an eye on the whole thing (has to run about an hour, usually), as you don’t want to lose small fragments. They will just run out of the gel if you wait for too long.
When the gel is finished, you drench it in ethidium bromide or something similar toxic. That stuff binds to DNA and is visible under UV light. That looks a bit like this:
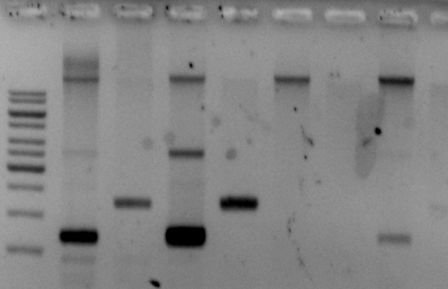
With the help of this, you can now determine several things: Did my PCR work? Did my digestion (= cutting DNA into specific pieces by using enzymes) work? Did process that produces a certain kind of DNA work?
You can even cut out pieces of the gel and extract the DNA!
Sometimes, a PCR is not the ideal way to create the product you want. Sometimes, actually, pretty often, you need to fall back on bacteria. And for that, you need plasmids.
What is a plasmid?
Generally, a round piece of DNA. Bacteria can take in these pieces and use them to, for example, become resistant to antibiotics if the DNA is coding for this specific ability. And as with most useful tricks from nature, scientists found a way to adapt this too.
It’s relatively easy to create a plasmid with the DNA you want. In addition to it, you need some other things I’m not going to mention because eh, you won’t remember anyway. The only thing I AM mentioning is the marker. The marker is usually a gene that is “visible” in a way, as the gene we want to insert doesn’t always have a visible effect on the bacteria that contain it.
Commonly used is a gene that provides resistance to a very common antibiotic.
I know, I know. We don’t want antibiotic resistance. That’s exactly the reason why it’s important to handle these kinds of organisms with a lot of care.
Anyway, the bacteria without plasmid are combined with the plasmid and then forced to take up this artificial DNA (mostly either by using a heat shock or by technically electrocuting them). They are then left to grow on, for example, a plate of medium that contains the antibiotic mentioned before. Those that contain the plasmid survive. Those that don’t … don’t.
The surviving bacteria are then put into liquid medium, so they can grow a lot. Then, they’re taken out of their broth, centrifuged a lot and broken open, to isolate the plasmids. Because while the cells were growing and replicating, they replicated the plasmid too and now we have A SHITTON of the plasmid.
And then? We can use the plasmid for stuff. Like a PCR.
Last one! If you’ve read the post about the building blocks of life I linked earlier, you should know that there is not only DNA but also RNA.
In biology, the usual way (it can happen differently too) is:
DNA -> RNA -> Protein
The step between DNA and RNA is called transcription, as the information on the DNA is transcribed into RNA to be translated into proteins.
Easy peasy.
In vitro transcription is exactly that, just inside a test tube instead of a cell.
That …. That’s it. You just combine a bunch of colorless liquids, put it in a warm place, wait for two hours and then put a tiny bit of it on a gel for gel electrophoresis to see if it worked.
If you still have questions (you probably do, this was just a really rough summary), feel free to ask in the comments. I will soon post my results so far, but they will only make sense if you’ve at least somewhat understood the methods.
Read some more about this:
Previously:
All about the Sperm-DNA - Vol. 1
Gene Editing - Cre/loxp - Vol. 2

Are you a microbiologist or a biochemist?
Neither, I'm an aspiring molecular biologist (= genetics).
I may never read or understand this, but you're like a smart person. Similar to but different from the so called precedent of the united states.
Dear @suesa, I finished reading the article and I feel there's a need to underscore something essential, when you talk about selecting for the bacteria having incorporated the right plasmid.
Bacteria are lean and mean energy optimizing machines - they compete to make the most of the resources available in their environment. When two bacterias try to reproduce, one with your plasmid and the anti-ampicilin gene inside and the other without, what happens:
After a few generation, the proportion of bacteria without the plasmid (without the anti-ampicilin gene) will be much higher. Those carrying the anti-ampicilin gene will be adversely selected (when there's no ampicilin in the medium). They'll start shedding the plasmid in order to compete on a level playing field with their brethren.
This is an important mechanism that needs to be understood by people throwing a fit over Genetically Modified Organisms. When the modification concerns the addition of a gene that confers a benefit under a selection agent then the risk of that modification spreading in the wild population is much much smaller than what the anti-GMO groups would have people believe.
Because absent that selection agent ( a proprietary pesticide for instance that the evil Monsanto wants to sell) having that genetic modification is actually a handicap for the organism.
And that is precisely Monsanto's business model: they do not want farmers to be able to maintain the modified varieties without also purchasing their pricy pesticide. So those modified varieties are usually quite fragile and impaired when let in a field without the pesticide .
This is to say that one of the most touted risks of GMO, that they might spread and "contaminate" the general population is, in many cases at least, the result of poor scientific reasoning: those genetic modification are ONLY beneficial in conjunction with some artificial selection conditions and are DETRIMENTAL absent those conditions.
In the wild, most GMO cannot compete with the non-GMO versions (if you don't also buy and spray the Monsanto poison)
Coming back to your post, this effect is seen in the fact that ampicilin is still used as an antibiotic despite generations of biology students unknowingly smearing anti-ampicilin bearing plasmids all over the place: the gene conferring ampicilin resistance is pretty big and costily to replicate and when there's no ampicilin around for it to be useful, bacterias quickly dump the resistance gene because it's slowing them down too much.
Students are certainly being super-careful but there's so many of them all over the world and mistakes happen from time to time. What "saves" us here is rather natural selection mechanisms.
Hi @suesa. If I might, I believe you do not do justice to the miracle of PCR and I would like to shed some light on several aspects that deserve a more thorough appreciation.
First you say:
Denaturation happens as you point out at 94-96°C That is a pretty high temperature that most biological molecules do not survive intact. The fact that single stranded DNA withstands such temperatures are testimony to its amazing stability.
The accuracy with which annealing happens I also consider quite amazing. Not something to be taken with a shrug, IMO
This is the most amazing thing of all. Most enzymes from normal organisms are designed to work at temperatures of up to 43 - 45°. Around 65+ most proteins (enzymes are proteins) start becoming denaturated. This is what happens to albumine when you cook an egg for instance.
Yet here you have a DNA polymerase that is happily replicating DNA at ... 72° ! Isn't that amazing ? It would be like if suddenly your "Spiegelei" turned into a live chicken and sprang out of your plate ! What's the secret ?
Well, that isn't just any vanilla-DNA polymerase that you are using there ! That is an enzyme which has been isolated from a species of bacteria adapted to living and multiplying near hot water sources like this one:
Thanks for the memories (my M.Sc in Mol. Biol dates from 1995 ...)
I was breaking it down as simple as possible, but that's why I linked something for further reading.
Btw, It's pretty rude to upvote your own comment but not the post you're commenting on. Just saying.
You're right. In my defense, I wrote that at about 3 am, right before crashing to bed, didn't even had to power to finish reading. Must've thought that I can come back later to finish reading. Corrected the rudeness :-)
I think even I followed that (for some reason I never really read/studied much about biology). I assume - and excuse my ignorance here - is this similar to the methods used in forensics to match DNA samples?
Actually, yes!
Biology made easy. This should be understood even by the dumbest of student and i think you @suesa should consider a career as a teacher. Awesome stuff.
I could never be a teacher, I hate explaining stuff over and over and over and over again to people who don't care at all. shudders
You dont really need to explain over and over and over again. Your explanations are the simplest i have seen and doesnt require much effort to grab. You might not take me serious but i strongly feel you wil make a great teacher.
Well, maybe not over and over again to the same people. But year after year to new classes of students. Idk. Not really my thing. I'm also too impatient.
But thanks for the compliment :)
You are welcome.
Nice job! A few things to remember:
PCR tips:
https://theupturnedmicroscope.com/tag/pcr/
https://goo.gl/images/2UQYW3
You can actually immerse polymerized gel with separated DNA into EtBr solution and it will intercalate well? We use to add EtBr in hot, liquid gel before polymerization and then inevitably inhaled cancerous stuff...
Our lab wants to cut down on EtBr usage so they are doing it like this. We have a basin with 1l water and a few drops of EtBr (so that the water is pink). The gel is then put into that for 15 minutes (10 for RNA). Then for a moment put in a basin with pure water (to get rid of leftover EtBr, we don't want it everywhere). Then you can put it under the UV light. Takes a bit longer but cuts down the toxic stuff you're exposed to!
It's a meme, who knows the source, but it's still great
That was printed out and pinned above my desk during my internship in the microbiology lab. Seems like everyone in this profession has the same humor :'D
This is simply iconic :) And "don't look at the laser with the remaining eye" in all spectroscopy labs (*even those without the lasers)
Didn't know that one but it made me laugh
Physicochemical classics :)
I remember by biochemist friend who was always talking about electrophoresis all the time.. Couldn't seem to grab what he was saying. But today i have learnt what it means from this simple explanations here
Thanks suesa.
Your friend should have just explained it, gel electrophoresis is first semester stuff. But I'm glad you understood it now :)
Thank you. I guess i never sat him down to explain it to me.