I am officially a doctor of biochemistry
Hello Steem community,
I have been absent for the last couple months because I've been busy finalizing my dissertation. If anyone is interested in taking a look, here is a link to download the PDF version... https://repository.lib.ncsu.edu/bitstream/handle/1840.20/35177/etd.pdf?sequence=1&isAllowed=y
...I thought I would provide a brief summary here. I worked on a family of enzymes called caspases. They are involved in regulating a programmed cell death phenotype called apoptosis (ah-puh-toe-sis). Everyday, your body produces 10 billion new cells, so 10 billion cell must be removed to maintain cellular homeostasis. Dysregulation of caspases leads to evasion of cells death, which is a hallmark of cancer; and too much cell death is a symptom of degenerative disorders like Alzheimer's disease.
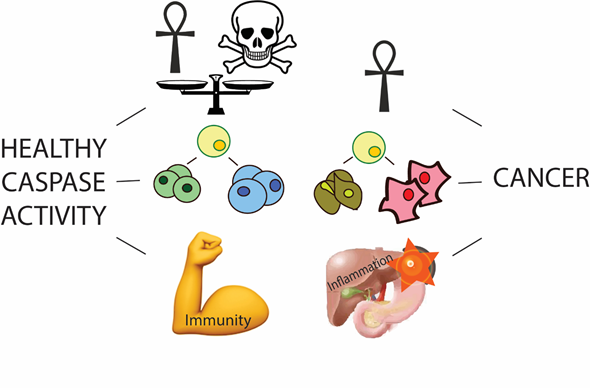
Caspase enzymes initiate apoptosis by cutting other proteins that maintain cell viability. There are 11 human capases that have evolved unique roles within the cell, and the structure of their active-site is evolutionarily conserved from their common ancestor. Due to similarity in the active-site, drugs that target the active site inhibit more than one caspase, so to develop a clinically efficacious drug, we need to learn about the naturally evolved mechanisms that regulate caspase activity.
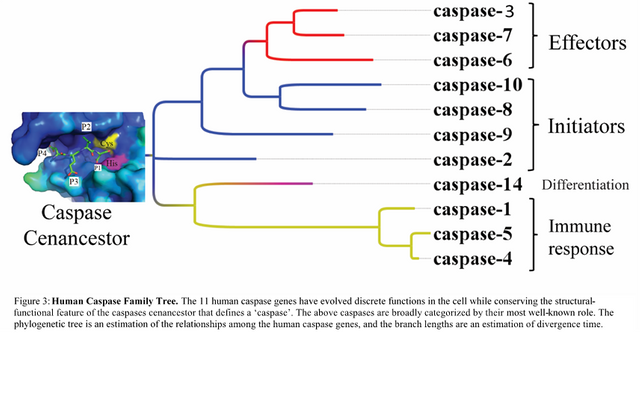
Only the effector and initiator caspases are involved in apoptosis as shown in figure 2, and my project focused on the effector caspases, also known as executioners. Interestingly, most cancer therapies are aimed at initiating the terminal executioners of apoptosis. For example, chemotherapy induces apoptosis via accumulation of DNA damage.
Caspase-3 and caspase-6 are relatively closely related, but they have evolved unique targets within the cell, so I set out to understand how the function of these proteins diverged over time. I used a phylogenetic method called ancestral state reconstruction, which has only been made possible since the advances in next-gen genome sequencing technology. I curated a database of caspase protein sequence and made it available at caspbase.org, and used the caspbase to construct several phylogenetic trees.
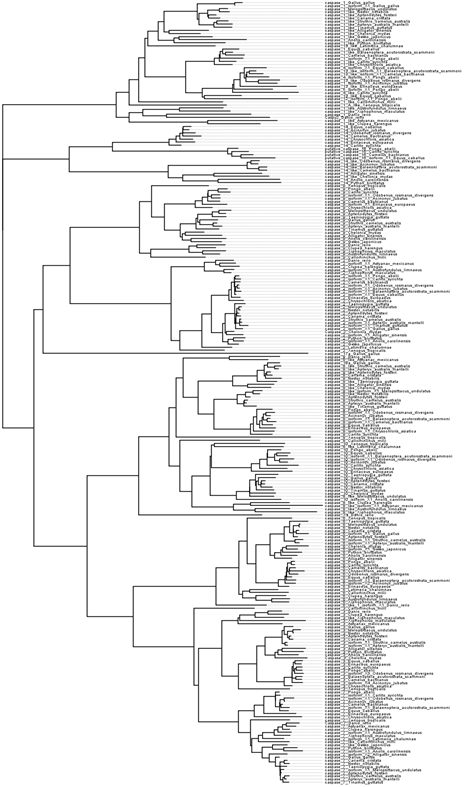
The nodes of a phylogenetic tree represent the common ancestor of the taxa on those branches. Ancestral state reconstruction uses a posterior probability distribution to predict the most likely amino acid sequence at each node. I then synthesized the reconstructed DNA, and resurrected several ancient genes in my lab. I could then test the behavior of the common ancestor of caspase-3 and caspase-6. Interetingly, the common ancestor (Node166) displayed similar behavior to both caspase-3 and caspase-6, which implies that they subfunctionalized after gene duplication. I also managed to solve a crystal model of Node166, and it is shown superimposed to models of caspase-3 and -6.
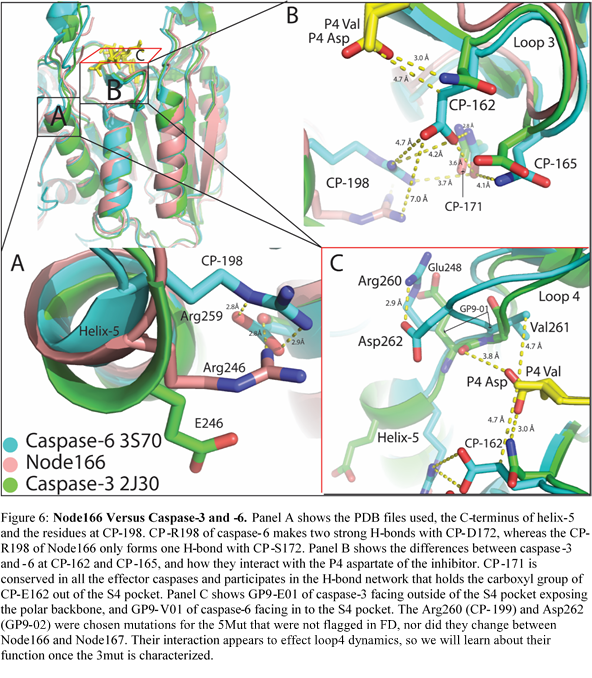
I discovered an interaction network of hydrogen bonds that stabilize an important amino acid in the active-site of caspase-6. The network was evolutionarily lost in caspase-3. These results change the current understanding of caspase substrate specificty. To investigate further, I made a series of mutations to Node166 that builds in the interaction network observed in caspase-6, and I managed to shift the substrate specificty of Node166 to reflect the behavior of caspase-6. An understanding of evolutionary sequence-to-function relationships will be critical in future drug development endeavors.
Much more detail and references can be found in the dissertation linked above. All the images were made by myself. Thank you for reading, and please ask questions if you are interested.
-TheScientist
Congratulations, and welcome to the club!
Congratulations @thescientist! You received a personal award!
Click here to view your Board of Honor
Congratulations @thescientist! You received a personal award!
You can view your badges on your Steem Board and compare to others on the Steem Ranking
Vote for @Steemitboard as a witness to get one more award and increased upvotes!