Determining the Gene responsible for providing the antimicrobial property from the Bacterial Genome
Hello Everyone,
In continuation to my post, where I explained the isolation and purification of the antimicrobial peptides. Now you must be aware of the Antimicrobial Peptides (AMPs) and Bacteriocins, if you have missed my last article you can find it here. Today, I’ll be sharing how to find the gene responsible for the production of these proteins by both the traditional way and the advanced way. As we all know about the central dogma where DNA converts to RNA and then to protein. All amino acids are been coded by three nucleotides (the triplet) and the resultant proteins gives the numerous properties to the living cell. So, the expression of a protein is based on gene sequence.
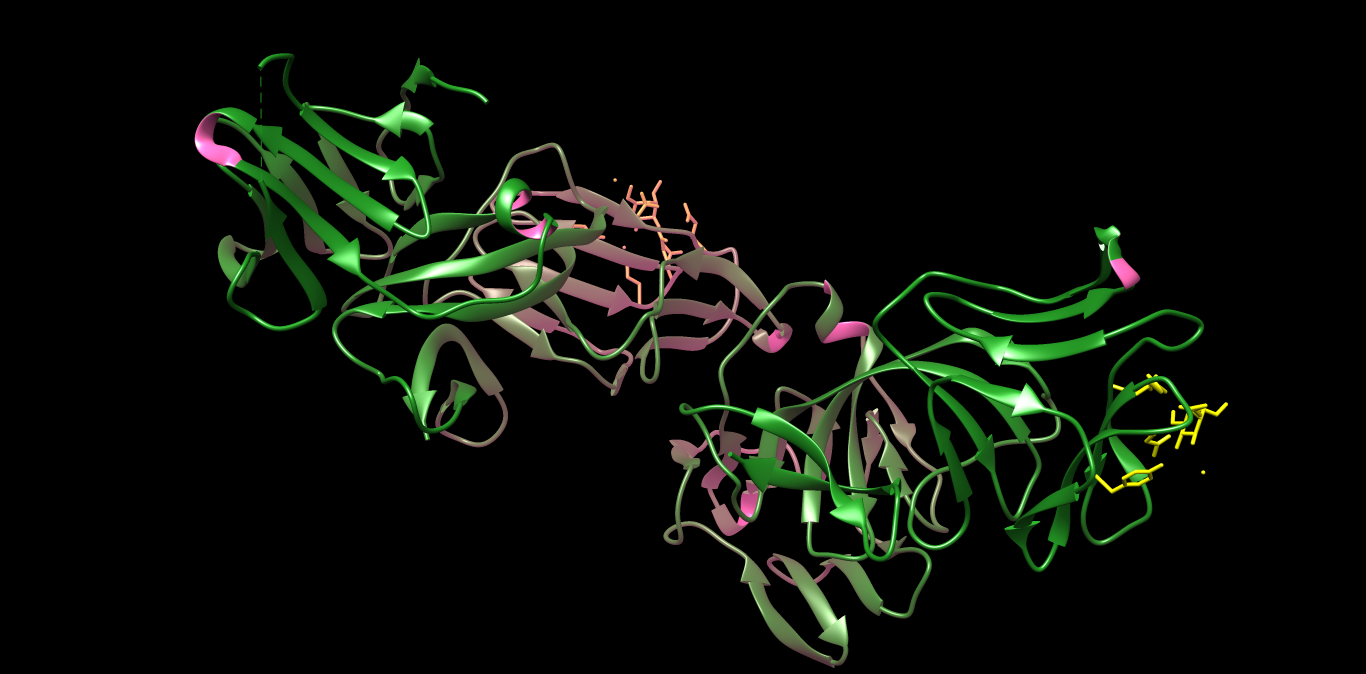
PDB 3MJ7 Structure made in UCSF Chimera 1.10.1 (Crystal structure of Bacteriocin from Pseudomonas sp.)
As these peptides are produced as the secondary metabolites, they can be extracted after the culture enters into the stationary phase of its growth. In brief, the peptide which was produced by bacteria are extracted from the culture and then processed for several purification steps and then was checked on SDS-PAGE for the size and purity, from there it was given for N-Terminal sequencing which gives the amino acid sequence of the peptide. It’s the old and traditional way of determining the sequence of peptide. From the sequence derived through the sequencing, universal forward and backward primers can be made and can be used to amplify the gene sequence from the whole genome of the bacterial species.
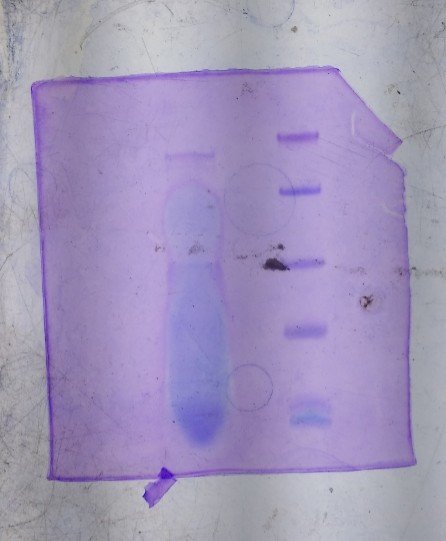 SDS-PAGE gel showing the AMP of 2.6KDa peptide
SDS-PAGE gel showing the AMP of 2.6KDa peptideSequencing of the purified peptide
Protein or peptide sequencing is a process of determining the amino acid sequence in the region you are interested in. Edman degradation and mass spectrometry are the two methods used to sequence the peptide, Mass spectrometry remains the most frequently used method for the sequencing purpose of peptide which determines the whole sequence of a protein. Whereas the Edman degradation includes the removal of a single amino acid at a time through Edman reagent and hence determines the sequence in that manner and are mostly used to characterize the N-terminal of the protein.
To start with, the composition of the amino acid in the peptide sequence must be known or at least must have an idea of the composition of amino acid. Because during the hydrolysis type of protease should be used to cleave the protein must be known. The whole protein was hydrolysed by 6M hydrochloric acid at 110°C for overnight or more which gives the small units of peptide with their constituent amino acids. The hydrophobic and charged amino acid are separated through the ion exchange chromatography and their derivatives are resolved through the high performance liquid chromatography. N-Terminal sequencing is benefited over the whole protein sequencing by the mass spectrometry, by using N-terminal sequencing the sequestral order of the peptide can be known exactly. N-Terminal sequencing generally follows these steps:
- Labeling of the terminal amino acids.
- Hydrolyzing the protein or peptide.
- Determination of amino acid by comparing with the standards through chromatography.
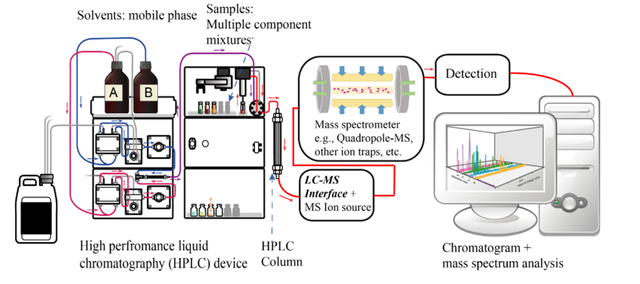
 HPLC system for the purification of Antimicrobial Peptides
HPLC system for the purification of Antimicrobial PeptidesIdentification of the peptide sequence by the Mass Spectrometry
Sequence of a part protein need to be determined for its nomenclature which was derived from the DNA sequence. The protein of interest is isolated through any of the chromatography techniques and then subjected to digestion as the protein involves with several regulatory elements and different subgroups with different secondary structures. The digested protein was first desalted to remove all the ion present in it through the dialysis (semi-permiable membrane) and then subjected to the MALDI-TOF (Matrix Assisted Laser Desorption/Ionization- Time of Flight). Alternative method of this is to desalt the peptide and separate it through the reverse-phase HPLC then analyze it into the mass spectrometry via ESI source (LC-ESI-MS) Liquid Chromatography-Electron spray Ionization-Mass spectrometry.After the sequencing of the peptide/protein the DNA/RNA sequence can be predicted or analyzed through the expasy.tools (SwissProt). This will give the overall idea of the nucleotide sequence responsible for the peptide formation.
More Advanced way of determining the Nucleotide sequence responsible for the peptide formation
Whole genome sequencing of the bacterial genome is the best way of determining the sequence. The only demerit of using whole genome sequencing is the money involve in it, but with the whole genome sequence you will end up with all the information you need to know about the bacterial species not only the AMP sequence but everything. If you see it from a student point of view you can also announce the genome of that bacteria which can also give you citation if someone want to use the genome of that bacteria. There are lots of bioinformatics tools available online which can help identify the sequence for the AMPs.

Wikimedia Commons Illumina HiSeq 2500
The tools blast your sequence file with the database and will give you the region having the NRPS clusters which generally carries the antimicrobial peptide in the upstream. Different families of these antimicrobial peptides contains different amino acids sequences rich in charged amino acids, antimicrobials peptides are generally known to have cysteine rich sequences. As the cysteine molecule forms the disulfide bond between the two amino acids and give the whole molecule a more compact and more stable structure. By these two methods you will get to know the sequence of your protein of interest, now the question come what is the need to do all that? Why we were doing all that degradation and sequencing.
Advantages of determining the peptide sequence
Determining the gene sequence will gives us the power to know and understand the importance of a single amino acid and single nucleotide in the sequence. Which amino acid is the key element to give the structural activity and which one is giving it the antimicrobial activity. Not only the peptide with antimicrobial activity can be sequenced any of them can be sequenced and can be added to the literature for its further use. By determining the sequence of the peptide it will be much easier to isolate the gene directly from the genome of different species of that genera as the universal primer can prime that stretch. Molecular biological studies can also be implied by knowing the sequence of these peptide, may be in my next coming articles I’ll be sharing the use of these sequence in the field of Molecular biology. The compact structure formation of these peptide is really hard to replicate after knowing the sequence also. In the bacterial cell these peptide forms these structure which are still very difficult to do, the whole mechanism by the chaperons and all are bit difficult.There are several fields where these peptide shows their potential role in the form of either therapeutics by providing the antibacterial and antifungal effect or as a probiotic which generally used to balance the intestinal microbial flora and in the field of food technology as preservative or antibiotics.
Conclusion
Recent advances has made us to identify the sequence responsible for the antimicrobial activity from the whole genome of a bacterial species. Having a knowledge of the peptide or nucleotide sequence always give an advantage to the researchers to make a more modified and more effective peptide which can have the ability to vanish all pathogenic microbes and make life more healthy.
Images which are not source are taken by me and do not violates any copyright.
For most of the terms I have hyperlinked with the simple Wikipedia page, that will make you understand if not feel free to ask anything below in the comment section. I will be happy to have them and will try to resolve doubts if anyone have.
References
Zhao et. al., 2016 BMC Genomics 17;882.
Ulmschneider et. al., 2017 Biophys J. 113(1);73-81.
Desriac et. al., 2013 Mar Drugs 11(10);3632-3660.
Yeaman et. al., 2003 Pharmacological Review 55(1);27-55.
Tambadou et. al., 2014 FEMS letters 357;123-130.
Leontiadou et. al., 2006 J. Am. Chem. Soc., 128(37);12156-12161.
Yang et. al., 2014 Front Microbiol. 5:241.
Luo et. al., 2015 Appl. Environ. Microbiol. 81(1);422-431.
Winn et. al., 2016 Nat. Prod. Rep. 33;317-347.

Are you doing all the work from peptide purification to Mass Spectrometry to genome sequencing. If so, bravo!
Yeah, we isolate the peptide from bacterial source, purify them and do all the characterization needed for the peptide and checking their activity in the cancer cell lines also, some of the peptides show effective killing in cancer cells also.
One of the peptide from our lab shows killing of leishmania also.
And species whose 16SrRNA shows similarity % less than 96%, we go for whole genome. We are also trying to clone the peptide sequence and produce them in E.coli. This work is going on in Dr. Bob Hankock lab in University of British Columbia, Vancouver.