DNA TECHNOLOGY: Gene Therapy and Cystic Fibrosis
Cystic fibrosis (CF) is a genetic disease that is due to one defective allele (I’ve explained this in details in some of my past posts). About one person in 25 carries the defective allele, but they also carry the normal allele and do not suffer from the disease. The normal allele codes for a protein called cystic fibrosis transmembrane regulator (CFTR). This essential membrane protein in epithelial cells transports chloride ions out of the cells and into mucus. Normally, when chloride ions are secreted, sodium ions follow, and this decreases the water potential of the epithelial mucus. Water follows outwards by osmosis, making normal, watery mucus, which can be moved by the cilia (tiny hairs) lining the airways.
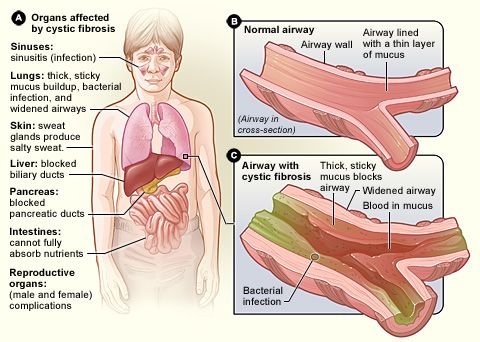
Cystic fibrosis sufferers make a protein that differs in just one of its 1480 amino acids. Although slight, this fault prevents CFTR from functioning normally. Chloride ions and sodium ions cannot be secreted, and so the mucus becomes much thicker than normal. Dead epithelial cells containing long DNA molecules also accumulate in the mucus, adding to the general congestion of the airways.
Sticky mucus is also a real problem in the pancreas. The mucus blocks the pancreatic duct, preventing the secretion of pancreatic juice. Treatment of CF includes regular physiotherapy in which the chest is massaged to dislodge the mucus. Even so, infections are common and most CF sufferers have to take a variety of antibiotics according to the infection they have at the time.
As cystic fibrosis is due to a defect in a single gene, it is a good candidate for gene therapy. In 1989, the cystic fibrosis allele was located on chromosome 7. The base sequence of the healthy gene was then compared to the defective allele, and the nature of the fault was narrowed down at the molecular level. This opened up the exciting possibility that if healthy genes could somehow be introduced into the epithelial cells, they might be expressed and so make the correct membrane protein, solving the problem for a time.
The basic steps are as follows:
- The CFTR gene is isolated, and cut out.
- The gene is cloned many times by PCR.
- The genes are encapsulated, by putting them into either liposomes (spheres made from lipid) or viruses.
- The gene particles are inhaled, so that they can pass into the epithelial cells of the lung to be incorporated into the DNA of the cells.
Once in place, if all goes well, the healthy genes are transcribed, making the correct protein.
Germline gene therapy and cystic fibrosis
Replacing the gene in body cells would still mean that the eggs or sperm of the sufferer would carry the defective alleles, which could be passed on to future generations. To prevent this, one future possibility is germline gene therapy. This is even more fraught with ethical problems as the original alleles in the zygote would be changed. The individual would grow and develop with healthy alleles in all cells and all of the individual’s children would inherit the healthy allele.
Germline gene therapy sounds attractive but there are problems, so much so this area of research is banned in the UK, and in many other countries. Ethically, there is the familiar ‘where do you stop?’ problem. A new treatment for cystic fibrosis or haemophilia would be desirable, but having the technology might allow for all sorts of temptations. Our knowledge of the human genome could reveal, for example, genes that code for intelligence, height or skin colour. Tampering with these characteristics would lead to obvious problems.
Biologically, germline gene therapy is potentially dangerous because we know very little about how genes function in the embryo. Tampering with genes in the zygote could have effects that might become apparent only in later life.

DOES GENE THERAPY EVER WORK?
The first gene therapy clinical trials that began in 1990 met with some success when SCID patients were treated. However, despite many other gene therapy trials since, there have been few other successes. As of 2007, no gene therapy treatment has been approved for use in human patients. Trials are still going on, but researchers are now more cautious. The field suffered a serious setback with the death of a patient during a trial in 1999. Confidence in the techniques was again shattered in 2003 when a gene therapy trial treating more SCID patients in the USA had to be stopped after a child in a similar French trial was found to have developed leukaemia as a result of the treatment.
Gene therapy has not achieved success for several reasons:
- It is difficult to get an introduced allele or gene to become stable in the host genome – as cells divide, they tend to lose it and patients have to undergo multiple rounds of gene therapy.
- Patients start mounting an immune response to the viral vectors, so repeated rounds of therapy become impossible.
- Viral vectors can be unpredictable when introduced into living cells – although treated to remove their ability to cause disease, they can revert.
- Most diseases, apart from the few genetic diseases caused by problems in one allele, involve several genes. The chances of being able to fix a one-gene problem are remote, fixing problems involving several genes looks impossible at the moment.
GENOMES AND GENE SEQUENCING
Sequencing individual genes or whole genomes involves discovering the exact order of the bases cytosine, guanine, adenine and thymine in the DNA strand or strands that make them up. The structure of DNA was worked out by Francis Crick and James Watson in the early 1950s, but it took about 30 years before researchers were able to develop a reliable sequencing method.
DIDEOXY METHOD OF GENE SEQUENCING
The most popular method for gene sequencing is called the dideoxy method, or Sanger method, and was invented by Frederick Sanger. He received his second Nobel Prize for this in 1980. The basic method is as follows:
The DNA to be sequenced is prepared as a single strand. The single strand of DNA then acts as a template to build up a piece of double-stranded DNA as new bases are added by the enzyme DNA polymerase I.
But, instead of doing this just with ordinary bases, the DNA is supplied with DNA polymerase I and two different mixtures of bases:
- A mixture of all four normal (deoxy) nucleotides in sample quantities: dATP, dGTP, dCTP, dTTP.
- A mixture of all four dideoxy nucleotides, each present in limiting quantities and each labelled with a tag that fluoresces a different colour: ddATP, ddGTP, ddCTP, ddTTP.
Because all four normal nucleotides are present, the second strand of DNA is built until, by chance, the DNA polymerase inserts a dideoxy nucleotide (one of the bases with the coloured tag). The polymerase enzyme is then blocked, as it can’t handle a base with this shape. It no longer adds any more normal bases – the process of making double-stranded DNA stops dead, leaving the strand unfinished. If the ratio of normal nucleotide to the dideoxy version is high enough, some DNA strands will become quite long before insertion of the dideoxy version stops the process.
Finally, the end result is a mixture of strands of double-stranded DNA, all of different lengths. All possible lengths are represented and analysis using gel electrophoresis separates out each length. By looking at which coloured at the end of each length, the sequencer can work out the sequence of bases in the DNA.
This is only one method of sequencing and, today, the whole process is automated and very fast, which is why so many gene sequences and whole genome sequences have been worked out in the past few years.
THE HUMAN GENOME PROJECT
A project to completely sequence the entire human genome began in the 1990s and is still yielding important results. Work on the project took 13 years and did not proceed without setbacks and controversy. A draft sequence was released in 2000, and this was improved and released as the full sequence in 2003. This revealed 20 000 to 25 000 human genes as well as the regions controlling them. The resulting DNA sequence maps are now being used as tools to explore human biology.
The DNA sequences announced in 2003 were only rough drafts for each human chromosome, like a sketch that is later built up into a detailed painting. Researchers are now concentrating on adding the detail. The final human chromosome to be sequenced in much more detail, chromosome 1, was finished in 2006. A project started in the same year to produce a map of human allele variants, which will be really useful for research into human disease.
Human conflict and the human genome map
The image of a scientist as an unfeeling and slightly weird boffin is a common stereotype, but scientists are very human, as the background to the enormously successful project to sequence the human genome demonstrates. The draft human genome, the sequence of most of the human chromosomes, was announced jointly in June 2000 by the Human Genome Project (HGP) a publicly funded international programme, and Celera Genomics, a US private company co-founded by Applera Corp. and Dr Craig Venter. Simultaneous publication of the research behind the announcement followed in February 2001. This public show of togetherness masked a bitter dispute that rumbled on for years.
The two groups used slightly different techniques to read the 3.2 billion base pairs of DNA that make up the human genome. The HGP method broke the genome up into manageable segments that were mapped before sequencing took place. Celera took a more sledgehammer approach: it broke the genome into tiny pieces and then used massive computing power to reassemble the read fragments. The HGP camp were put on the defensive because Celera managed to produce a draft sequence in a fraction of the 10 years it took the HGP scientists, not to mention all the public money that they spent. Celera were criticised for the quality of their work, and for ‘cribbing’ essential information from the publicly available HGP work, which was updated as work progressed. Many scientists hoped that the joint announcement of the results would be the end of the rows, but a subsequent paper in a leading medical journal contained an article from HGP scientists, again criticising the quality of the Celera sequence.
LOOKING TO THE FUTURE
As well as adding more detail, the story of the human genome is far from over. The genome sequence is a bit like a map – it’s getting more detailed, but the really interesting part will be the journeys that we will be able to take using that map. Other genomes from different mammals, lower animals and prokaryotes have also been sequenced and vast projects are underway to see how entire genomes are related. Doing this will enable us to compare sets of expressed RNAs or proteins, gene families from a large number of species, variation among individuals, and the classes of genes that regulate the expression of other genes.

In the present decade we may be able to find:
- the exact locations, sequences and expression of individual genes;
- the mechanisms by which genes are regulated;
- exactly how the DNA sequence is organised into genes – where are the non-coding sequences and what do they do?
- the detailed structure and organisation of chromosomes and the function of proteins associated with DNA in chromosomes;
- how gene expression, protein synthesis and post-translational events are controlled;
- more about why some genes have been conserved in different organisms throughout millions of years of evolution;
- how SNPs (single-base DNA variations among individuals) correlate with health and disease;
- if we can tell from the variations seen between gene sequences in different individuals whether or not they will develop particular diseases;
- which genes are involved in complex traits and multigene diseases:
- more about how genes interact in the development of a human being from a fertilised egg.
I will like to pause here for now. In my next post, I will discuss more on organism cloning, and the technical and ethical problems with cloning organisms.
Thanks for reading.
REFERENCES
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3681190/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3681190/
https://study.com/academy/lesson/germline-therapy-definition-procedure-ethics.html
https://www.cell.com/molecular-therapy-family/molecular-therapy/fulltext/S1525-0016(01)90420-4
https://www.sciencedirect.com/topics/medicine-and-dentistry/germ-line-gene-therapy
https://www.yourgenome.org/debates/is-germline-gene-therapy-ethical
https://ghr.nlm.nih.gov/primer/therapy/genetherapy
https://ghr.nlm.nih.gov/primer/therapy/procedures
https://www.scientificamerican.com/article/experts-gene-therapy/
https://learn.genetics.utah.edu/content/genetherapy/success/
http://www.nature.com/scitable/topicpage/dna-sequencing-technologies-key-to-the-human-828
https://en.wikipedia.org/wiki/Genome
https://en.wikipedia.org/wiki/Whole_genome_sequencing
https://www.sciencedirect.com/topics/medicine-and-dentistry/genome-sequencing
https://en.wikipedia.org/wiki/Sanger_sequencing
https://www.ncbi.nlm.nih.gov/pubmed/18265267
https://en.wikipedia.org/wiki/Sanger_sequencing
It's a wonderful article and stands well on its own...in the eyes of this lay reader. I was impressed by your discussion of the ethical considerations and also by the backstory you provide about Celera and HGP.
I have a rudimentary understanding of genetics--derived from a college bio course and casual reading. However, I was able to understand everything in your piece. I also appreciate how you explain the difficulty in CF with mucous and repeated infections. I didn't realize that the mucous also affects the pancreas.
All in all, an enlightening article (for me). Thank you!
Sharing on Twitter :)
Thank you, @agmoore2.
Hello,
Your post has been manually curated by a @stem.curate curator.
We are dedicated to supporting great content, like yours on the STEMGeeks tribe.
Please join us on discord.
Good effort here, no doubt. It would have been better to link back your previous articles on genetic disease/defective alleles though.
Thanks, @gentleshaid. Here's the link.
This post has been voted on by the SteemSTEM curation team and voting trail. It is elligible for support from @curie and @minnowbooster.
If you appreciate the work we are doing, then consider supporting our witness @stem.witness. Additional witness support to the curie witness would be appreciated as well.
For additional information please join us on the SteemSTEM discord and to get to know the rest of the community!
Thanks for having used the steemstem.io app and included @steemstem in the list of beneficiaries of this post. This granted you a stronger support from SteemSTEM.
Have a look at $EDNA, its an project on EOS, you like it base on this article your wrote here
Thanks, @daudimitch. I'll check later.
Just like to add a note: There is obvious interest in this--so far I've had 3 click-throughs on Twitter. That's not typical.
Thanks once again.
Congratulations @loveforlove! You have completed the following achievement on the Steem blockchain and have been rewarded with new badge(s) :
You can view your badges on your Steem Board and compare to others on the Steem Ranking
If you no longer want to receive notifications, reply to this comment with the word
STOP