Sciencepic of the day: Next-Next generation sequencing
In todays post, i will share an image that was taken today in my group. It is a pic of a computer screen (oh, how boring!!). What is it showcasing though? what do these color bars represent?
Firstly, I should say what the computer is hooked onto. The computer is connected to a MinIOn sequencing device (errrmmm a what?). Its one of the latest nucleotide sequencing techniques out there, developed by the company Oxford Nanopore.
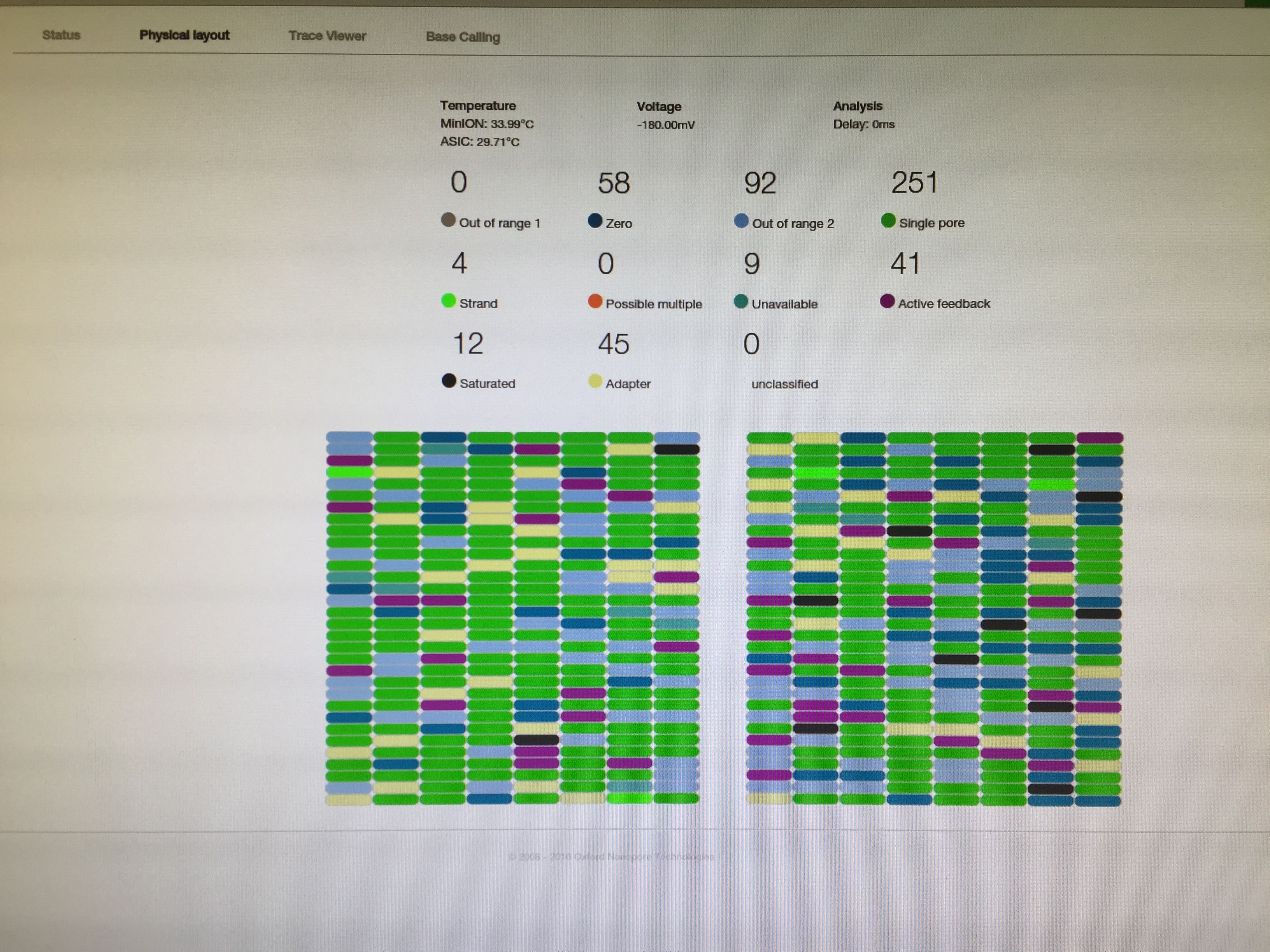
The image. A computer screen displaying a physical map of nanopores that are engaged in sequencing of a DNA sample. The color represents the status of each pore and the base that travels through it. This is a static image, if you were to film it, you would see the colors change constantly. Source: Own collection
Ok, so what does it do then?
Good question. In molecular biology, we rely on the availability of sequences that specify the structure, composition and functions of the molecules we study. Without those sequences, molecular biology would not exist or practicing in this field would become impossible. We therefore like to sequence the organisms we study. Initially and in the early days, this meant sequencing gene after gene, basepair by basepair. Technological advantages however have allowed us to scale this up significantly, allowing us to sequence complete genomes by generating short sequences (billions) that are assembled into one or more theoretical sequences. Rather then sequencing billions of short fragments, this device allows us to sequence a smaller number of larger molecules, circumventing this massive assembly challenge.
How is it different from other sequencing technologies?
Firstly, the chemistry and approach are completely different. Classical and next generation sequencing approaches all rely on the synthesis of DNA molecules and incorporation of modified nucleotides (each nucleotide having a different color). Incorporation of these bases allows very sensitive detectors to identify each incorporated base and read the sequence in this way. This technology is different as it does not rely on synthesis and incorporation of nucleotides. Rather, small pores (nanopores) made of proteins, are employed in the MinIon device that capture single DNA molecules and pass them through the pore. Whilst the DNA molecule travels through a charged pore, each base that passes can give a specific electric signature signal, which sensors can pick up and use to generate a base signal (also called a squiggle). These are then analyzed to determine what the identity of each base is and generate a sequence (base calling). Since there is not a synthesis requirement, very long molecules can be guided through these pores, leading to very long sequences that do not need to be assembled too much (theoretically). It also means that native DNA and RNA molecules can be sequenced including their possible modification (for example, see Simpson et al., 2017), something that other technologies cannot do.
Figure 1. Nanopore MinIon device. Source
As with most technologies, there are limitations. At this stage in the game, the accuracy of the device needs improvement (it makes base calling errors), but in combination with other more accurate sequencing technologies, this device allows rapid sequencing of full bacterial and eukaryote genomes as described in Loman et al., 2015; Goodwin et al., 2015; Debladis et al., 2017). There is little question though that this technology will change the face of biology. Accessibility to this device is far greater (i have it in my laboratory) then those super expensive Illumina or PacBio sequencers. Allowing biologists to conduct their own genome sequencing and analyses projects in house.
Research in Molecular Biology becomes ever more exciting (and challenging if you wish to keep up!!)
REFERENCES:
Debladis, E., Llauro, C., Carpentier, M. C., Mirouze, M., & Panaud, O. (2017). Detection of active transposable elements in Arabidopsis thaliana using Oxford Nanopore Sequencing technology. BMC genomics, 18(1), 537.
Goodwin, S., Gurtowski, J., Ethe-Sayers, S., Deshpande, P., Schatz, M. C., & McCombie, W. R. (2015). Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome research, 25(11), 1750-1756.
Loman, N. J., Quick, J., & Simpson, J. T. (2015). A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature methods, 12(8), 733-735.
Simpson, J. T., Workman, R. E., Zuzarte, P. C., David, M., Dursi, L. J., & Timp, W. (2017). Detecting DNA cytosine methylation using nanopore sequencing. nature methods, 14(4), 407-410.
I hope you will find these pieces interesting. Your vote and follow help me open up the exciting world of scientific research to you. Thank you @foundation for this fantastic SteemSTEM gif

Nanopore MinIon in the field!
Thanks for sharing that.had not see. That one before. I hope to be out in the field sequencing pathogens at some point 😁
that is seriously impressive man ! amazing how far our technology has evolved !
I know. to be clear, I did not invent this! just a user of the technology. i agree it is amazing. the device is smaller then a phone, has a USB port that connects it to the computer (which needs to be fast) and off we go!
Its a crazy coincidence, but I'm using a minION sequencer today as well. I've been trying to get certain sample preps to work with the RAD-002 kit. I also wrote a post last month about the SmidgION, a portable version of the MinION.
Do you use the MinION a lot or is it just for infrequent sequencing jobs?
We have just started using it really. This is our second run. Straight genome sequencing for now. I will check out your post . Missed it!!
The minion isn't portible? It fits in your hand :)
Oh Oxford nanopore.
The MinIon still needs a decently powered laptop with a solid state hard drive to run while the SmidgION is supposed to run on a smartphone.
Ah, good points.
For it's size, the minion is a pretty amazing DANA sequencer. When we were using one in a lab I worked in, it made a lot of bad calls though. However this was years ago and I suspect the technology has improved by leaps and bounds since then.
It still may. I can update you on that later. This is only the second run. Amazing technology thoug