What's up with nucleases?

Depending on how much contact you have with me on steem.chat or discord, you know or don’t know that I’m currently working on my bachelor thesis. This caused my posting behavior to go from “once a day” to … well, “once every one or two weeks, maybe.”. A bit over a month and things should be back to normal, although I assume it’ll be hard to go back to the regular posting I did almost a year ago (my Steem birthday is 16th of June!).
Nevertheless, I want to provide you with some scientific content again. While writing the introduction to my thesis, I had to do some research on nucleases. After finishing that, I looked at all the sources cited and thought to myself ”Hey, I already put so much work into this, I could just as well use it to get some easy money on Steemit teach people something!”.
So, today, I’ll present to you a post about nucleases, the things that are used to edit genomes.
If you don’t know what DNA is made of, can’t tell the difference between amino acids and nucleobases, and think guanine is a fruit, please read my post on the building blocks of life for a fun introduction to … uhm, the building blocks of life. You’ll need it. I mean, you can read this post without having understood any of the things mentioned. I will include silly sketches. Maybe those help.

What’s a Nuclease?

After you’ve read the introduction to “the building blocks of life”, you now know that nucleobases are the things DNA consists of. The word nuclease consists of two parts: Nucle- and -ase. The nucle- refers to the nucleobases, the -ase is a word ending usually used for enzymes. Most of you probably think that enzymes only break stuff down (that’s what you’re usually taught), but in general, enzymes catalyze biochemical reactions that would take millions of years if left to their own. Pretty neat, right? @suesa
Nucleases specifically are enzymes that make changes to DNA or RNA, which both are made up of nucleobases.
Which explains why nucleases are used for genome editing! If you want to make changes to a genome, you use enzymes that catalyze such a reaction. But what does actually happen?
The general way nucleases used in genome editing work is that they introduce a break to the DNA double-strand. This break is then repaired through one of two pathways: Nonhomologous end-joining (NHEJ, I am not going to write this out every time) or homology-directed repair (HDR, not going to write this again either)1. What’s that? Well, it’s in the name!
NHEJ does not require a sequence homologous to the DNA that has been broken in two. Instead, enzymes simply “glue” the ends together in the best way they can. This can lead to random insertions or deletions (so-called “indels”, not to be confused with “incels”) of nucleobases.
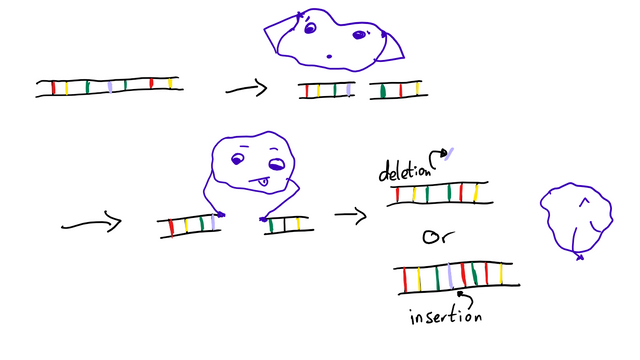
Cool way to deactivate a gene by causing a random mutation. Not so cool if you want to insert something specific. That’s where HDR comes in.
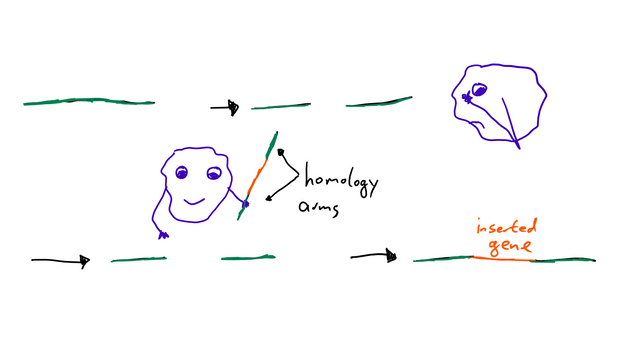
HDR is a repair of DNA directed by homology, which means the enzymes that repair the DNA have a template they use. A blueprint for what they want to achieve. In a natural context, HDR uses the second copy of the gene that’s available in the cell. But if we want to introduce a DNA sequence artificially, we need to provide that template ourselves. This template needs two “homology arms” (= sequences that are the same as the DNA sequence where the break happened) and between those, the insert (= piece of DNA we want to introduce). Then some biochemical magic process happens, and we got our mutation! Easy as that.2

ZFNs and TALENs

Biologists love their abbreviations. Then again, who wants to write a text about zinc finger nucleases (ZFNs) and transcription activator-like effector-based nucleases (TALENs) and write out the full name every damn time? Not me. Certainly not most of the others. So we make it easy and use an abbreviation. But what’s behind those? And why do I group two nucleases, that sound completely different, together? Am I that lazy?
Yes, but that’s not the reason I’m putting those two in the same section. ZFNs came first. They were discovered in the egg cells of a specific frog (Xenopus laevis, for those interested)3. What do they do?
ZFNs have, as the name suggests, zinc fingers. Those have a specific nucleobase triplet they target. 3-5 fingers per ZFN result in a pretty good recognition of a target sequence where the DNA should be broken in two. Even better, you need two ZFNs, which each has their set of “fingers”, to create this break.4 You can imagine it like this:
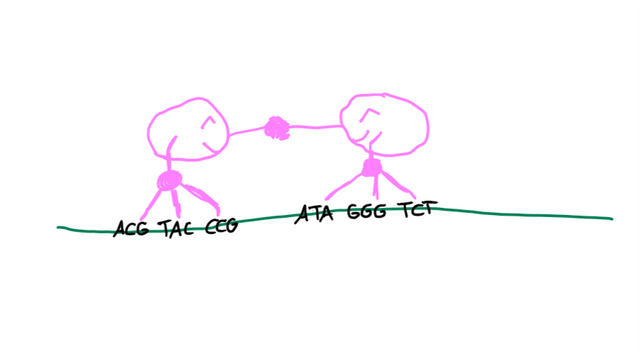
The two ZFNs find the DNA sequence, they dimerize (“two get together and connect”), they break the DNA, the DNA is repaired through either NHEJ or HDR, everyone is happy.
What about TALENs?
Basically, the same concept.
TALEs (transcription activator-like effectors, basically what I wrote earlier just without the “nuclease” part) are proteins from a certain kind of bacteria that infects plants5. Those alone can not break DNA. But they can find and bind to DNA sequences! And that very efficiently6!
Biologists don’t like to see good systems go to waste. And they already knew how ZFNs work. What they then did was taking a ZFN and replacing its zinc fingers with the system TALEs use for DNA recognition7. Who would have thought that real-life Frankenstein would be so … boring.
That’s the only difference between ZFNs and TALENs. That’s why I grouped them together. Understand one, understand the other! If it was just always this easy.

CRISPR/Cas9

CRISPR/Cas9 is the newest system using a nuclease to induce DNA breaks, and …
Oh, look at the time! I think CRISPR/Cas9 has to wait a bit more; this post is already very long.
Stay tuned!


Sources:
1Gaj, T., Gersbach, C. A., & Barbas III, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Cell Press, 31(7), 397-405. doi:10.1016/j.tibtech.2013.04.004
2 Resnick, M. A. (1976). The repair of double-strand breaks in DNA: A model involving recombination. Journal of Theoretical Biology, 59(1), 97-106. doi:10.1016/S0022-5193(76)80025-2
3 Miller, J., McLachlan, A. D., & Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO Journal, 4(6), 1609–1614.
4 Carroll, D. (2008). Zinc-finger Nucleases as Gene Therapy Agents. Gene Therapy, 15(22), 1463–1468. doi:10.1038/gt.2008.145
5 Bogdanove, A. J., Schornack, S., & Lahaye, T. (2010). TAL effectors: finding plant genes for disease and defense. Current Opinion in Plant Biology, 13(4), 394-401. doi:10.1016/j.pbi.2010.04.010
6 Schornack, S., Meyer, A., Römer, P., Jordan, T., & Lahaye, T. (2006). Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. Journal of Plant Physiology, 163(3), 256-272. doi:10.1016/j.jplph.2005.12.001
7 Christian, M., et al. (2010). Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics, 186(2), 757-761. doi:10.1534/genetics.110.120717

I think your article should be on "programable" nuclease as used in genome editing. As the ZFNs, TALEs, Cas9 all can be guided to a specific sequence by the researcher. Before these nucleases, the "bread and butter" of molecular biology were the restriction nuclease which really revolutionized molecular biology. These nucleases cut a specific sequence eg. EcoRI GAATTC or HindIII AAGCTT and still very useful today.
You're correct. But explaining that too would have made the post even longer. It's the usual problem I face on here when trying to write for a broad audience: Do I make it 100% correct, or do I gloss over some details to make it easier to digest? I usually take the second path.
Yes it is hard to be comprehensive but in a post about nucleases, someone who doesn't know what a nuclease might think of only the types you mentioned. So something like "What is a nuclease?" A nuclease is an enzyme that cuts nucleic acid (DNA or RNA) and scientist can direct nucleases to cut at specific sequences to edit genomes....(then mention the 3 as in your post). I just felt that there is so much history in the discovery of restriction enzymes that your post should have mentioned these very important nucleases. The Nobel Prize in Physiology or Medicine 1978 was awarded to Werner Arber, Daniel Nathans and Hamilton O. Smith for the discovery of restriction enzymes and their application in molecular biology.
Now you mentioned it :P
ha ha as long as people read the comments.
Best of both worlds: The easy to digest analysis on top and the nitty gritty historical details in the comments. Upvoted for visibility.
Great article! I skimmed it before reading it and got sad you had so little on CRISPR written. Definitely fix that in the next post! Im curious, what is your thesis actually about? (I just graduated in biochemistry so I'm interested.)
Also, why did you refer to the building blocks as nucleobases instead of nucleotides?
I'm definitely going to be joining your discord. I'd love to start writing more science and tech articles here considering my background. I just feel like I haven't really found my science community here yet.
Great post! I'm a big fan of anything that helps bring science to the masses!
Resteemed for sure.
Ah, I think I said "nucleobases" in my "Building blocks of life" post and didn't want to confuse people, so I stuck with it. But you're right, it's not completely correct.
Ofc I see this comment after inviting you to the discord server ^^
The topic of my thesis is basically that I try to combine CRISPR/Cas9 with Cre/loxP for more efficient gene knock-in.
If it works out that would really be great! I'm really excited to see where this tech is in five years. I think it's going to be integral in fixing so many diseases and disabilities! It's definitely going to be the greatest medical breakthrough in the 21st century so far once it is fully realized! Good luck on your thesis!
I have a guanine bush planted in my house. :P
It's good to read you again, now I understand why you disappear for so long.
I don't think it's easy money, it took you time and effort to acquire that knowledge! Greetings!
Well, yeah, it didn't took me super long to write this post, so it could be regarded as "easy money". Then again, researching all that in the first place ... took a while.
Sounds like we have similar interests. I am retired after a 40-year career in environmental biotechnology and biomedical research. I call my blog "Unpublished Data" because I'm going through all of my backups and assembling articles for publication on Steemit. I have many unfinished manuscripts and years of data which I want to share with fledgeling scientists in the hopes that my work will be appreciated and inspire further research. When you have time, would you say a few words about how I can better use steemchat and discord? I only have one contact who friended me, and I'm not sure how to engage other than posting and commenting. Thanks!
I think you'd benefit greatly from joining the steemSTEM discord (click the last image in my post). SteemSTEM is the science community around here, we'd be happy to have you!
I already have joined the SteemSTEM server on Discord, but I have only one contact (who gave me some advice when I asked a question). How do I gain more contacts? Also, what's up with Steemit Chat? It never loads or opens. Thanks!
Very detailed and well-explained post. The diagrams were cute. :)
I've a scientific topic for you and may be you can cover this:
Good luck @suesa with your thesis.
I'm putting it on the list! Certainly an interesting topic, but that will need some research, so I won't be able to write about it before turning my thesis in.
I'm glad that you are adding it to the list. Your thesis must be the first priority. I fully understand that.
If you are to busy writing your thesis, I would be more than happy to help out! I just finished my degree in biochemistry and have been trying to think of topics for a first science article here.
Thank you, but I prefer to write my posts myself ^^
I invite you to join the steemSTEM discord (click the image at the very end of my post) to connect with some fellow scientists!
Just joined this morning! I'm excited to have hopefully found a good community of people like me on here!
I see papers describing genome editing as a simple inject-nuclease-wait for embryos to grow thing. However, how does the nucleus enter, for example blastomere nuclei,
getting through the nucleolemma
reaching condensed solenoids that possess epigenetic modification and structural factors that prevent the binding of factors?
Well, the genome editing I personally have been doing for the past few weeks involves injecting the solution in a zygote before it reaches two-cell stage, so I don't have any insights on how it works in later stages :/
Oh, that's okay. thanks for the response though :)
First of all, yes. If you haven't read the other post, go there if you are to understand any of this.
These kinda of posts are the ones I read imagining myself as a biologist performing non-invasive surgery on self... mesmerized by the details in one body. In just a strand of DNA. Or a cell. How enzymes work 24/7 to play their roles just to keep my body functioning.
Lol. Too deep.
I love your posts. How you clarify things. Simplify them for those of us who are interested but can't go back to school for it :D Also for the fact that you have made it clear that we (your readers) should fall in love with your
awesomedrawings xDFinally... Ever thought of being a lecturer? I imagined you stating that your class should master 'biologists abbreviations' because you don't have the energy to write something like;
Or
Look at that, a lengthy reply too. Maybe I should end this but before I do, hopefully you've been good Suesa. All the best with your thesis :)
I couldn't be a lecturer, I have no patience with people who only attend a lecture because they have to xD
Most of our lecturers abbreviate everything though, you either learn what it means or you don't understand a word.
You are a Steemit lecture though xD And here we understand we have to carefully read the post to understand anything and that our lecturer hates spammers xD Thank you for what you do.
Cool to see you posting science again, and as always I really enjoyed your sketches.
DNA repair is of great relevance in toxicology aswell, as it's there when damages to the DNA are made permanent (of course the mechanism for single-strand repair are important too).
In the end, DNA repair might be the most important process in our bodies. After all, aging is tied to it too.
It's extremely important. Just with like anything important, errors here can have grave consequences.
Hello @suasa
Good to know that your steemiversary comes this June. Looking back and see how far you have come, gives reason to be grateful to those who have supported you thus far. It is my first time to know you're currently working on your thesis and yet putting in much effort to ensure this place remains a trusted destination for scientists seeking one information or the other. We are grateful to you.
This article on nuclease enzyme and its mechanisms of actions, is so detailed that is even a non scientist will get the message. I like the way you use those sketches to illustrate your points. It helps pass the message so well.
I am staying tuned for the concluding part of this piece in which you hope to treat CRISPR.
Regards,
@eurogee of @euronation
I've tried to buy a TV of HDR-quality but when I was going to buy one, I just read this post and now I want a ZFN-TV!
And another comment related to post - even though I don't personally accept everything, I have some friends who are homology too and I've heard it's not their choice but it's in their DNA.
So I'm cool with homology. Just as long they keep their arms out of my butt.
I'm so glad we're friends 😂