Journal of Unpublished Results – A case for the validity of scientific research (Part I)
Welcome Steemiacs, scientific affiliates and science-skeptics alike!
Preface:
In Steemit, we often witness and take part in fervid discussions on scientific topics. Frequently, the results of the debate seemingly turn scientific evidence into a matter of opinion.
Those, who oppose the sources and arguments provided by the proponents of scientific establishment, claim that they have done their own research themselves, and came to their own conclusions accusing the other side of not citing the proper science and, instead, propagating views that the pharmaceutical industry has paid them to, in order to maximize capitalistic profits.
My major concern with these discussion is that results of major scientific studies are questioned. I am relating to comments like “the whole idea of vaccination is based on wrong science” (or in a similar phrasing).
In this post I will not be judging if vaccination (MMR, HBV or pertussis, doesn't matter) is right or wrong.
I will just cautiously imply that understanding the established scientific views, that our mainstream health system is based on, does not take a few days or weeks of scouring google and the interwebs for relevant information.
It takes year-long theoretical and practical training, as well as monetary, logistic and instrumental resources to verify even the most basic scientific findings.

To prove that someone with these pre-requisites (i.e. Master's in Biomedical Science) can verify and validate a specific scientific discovery, independently from the original scientists, in a different environment and in another experimental system, without any monetary incentive or perspective, I will present you here a peak into some of the experimental work I have done in the previous years. I am comfortable publishing the results here, because, what I present, is nothing novel or surprising given the body of work already conducted on the topic and will not be published in any journal in the future. While doing that, I will omit some unimportant details and simplify the molecular and methodological aspects.
Introduction:
All multicellular organisms originate from a fertilized egg cell, also called zygote. The zygote starts dividing into progeny that contains embryonic stem cells. These pluripotent stem cells are able to produce all the two hundred or so different cell types in the human body during embryonic and fetal development.
The organization of our multicellular tissues is considered to be hierarchical, i.e. there are stem cells in each tissue that are bound to produce specialized cells of an organ. These specialized cells are called “differentiated” and maintain their cellular identity over their own life time until they die off (e.g. skin cells), or even over the whole life span of the organism (heart and brain cells).
This developmental hierarchy of cellular differentiation was depicted by Conrad Waddington in his famous epigenetic landscape (1):
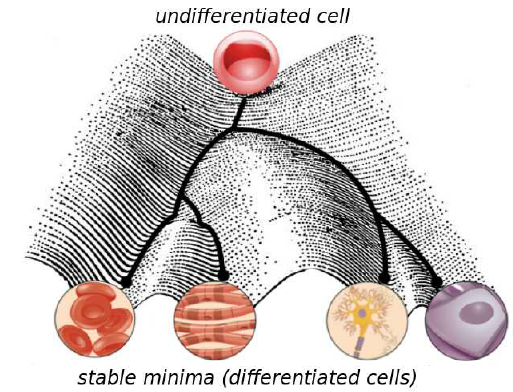
Beginning from the top (imagine embryonic stem cells), the activation and repression of certain genes decides upon the further fate of the cell. Generally speaking, cell progeny is like marbles rolling down the hills through one or another valley, ending up being one or another cell type. What is remarkable in this model, is that for a differentiated cell it is impossible to walk back on its path and become another differentiated cell. This is what the figure means to express by “stable minima”.
But is this truly the case? Is it not possible for a differentiated cell to “transdifferentiate”, i.e. change its cellular identity towards another cell type? Although it is still not known under which conditions and if such a process spontaneously happens in vivo, research from the last three decades demonstrated that it is, indeed, possible to transdifferentiate cells through genetic manipulation in the laboratory.
The first demonstration for this principle was provided by Weintraub and colleagues in 1987, who could transdifferentiate fibroblasts (connective tissue cells which produce collagens, e.g. beneath the surface of your skin) into muscle cells by the genetic introduction of the transcription factor MyoD (2).
That was pretty cool, kicked off research into cellular reprogramming and questioned the general validity of Waddington's diagram.
Let's switch gears a bit and look into cells of the immune system. You can read on some of its cellular components that I will mention in @suesa 's post and in @kryzsec 's post.
Here is a diagram of the hierarchy of the hematopoietic system that includes the immune system. As you can see, the hematopoietic stem cell, residing in the bone marrow, sets off its progeny to walk along several Waddington valleys for differentiation into several different cell types, including red blood cells and blood platelet producing megakaryocytes which we will not cover here further.
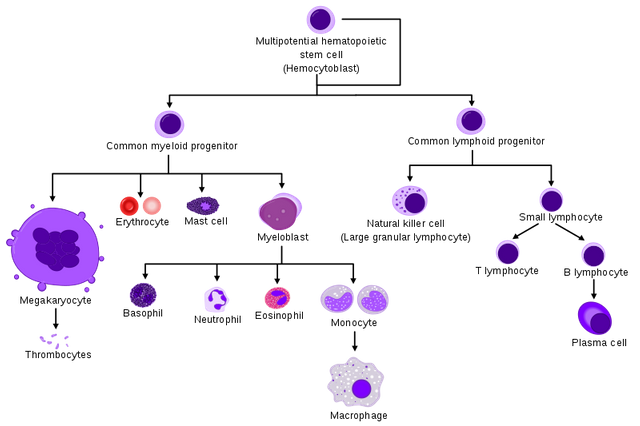
Instead we will focus onto the myeloid/monocyte (starting from myeloblast) and lymphocyte branches.
On the one hand, we have two important proteins/genes for the development of the myeloid phagocytic and granulocytic cells: the very related C/EBPα and C/EBPβ transcription factors. The importance of one of them is highlighted by the frequent mutations observed in C/EBPα in a subtype of acute myeloid leukemia (AML). The mutation blocks C/EBPα 's directive function to differentiate immature cells into granulocytes, thus leading to uncontrolled division of malignant blasts (sort of cancerous stem cells). (3)
What is this transcription factor thing I am talking about? It's a protein that directly binds to DNA and regulates the expression of a certain set of genes. Here's a picture of the molecular structure of C/EBPα (purple) binding to DNA.
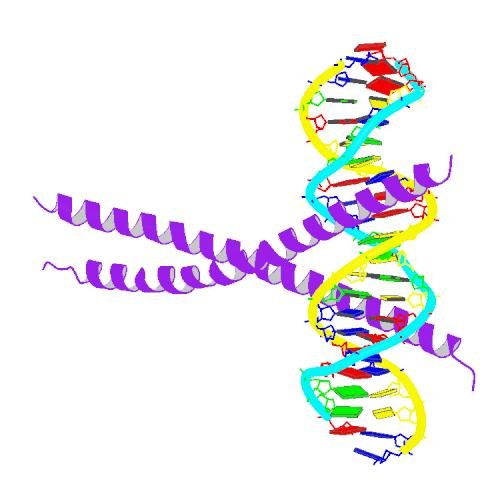
Can you see the DNA double helix :-)?
On the other hand, the transcription factor Pax5 is important for the development and maintenance of B-lineage lymphocytes. Mutations in Pax5 are often found in acute lymphoblastic leukemia (ALL), leading to expansion of malignant cell clones because of their inability to differentiate. (4)
It has remained silent for a long time in the reprogramming field since the initial characterization of MyoD's reprogramming capabilities. Then, in 2004, a publication by Thomas Graf and his team appeared in the journal Cell that announced the dawn to a new era of cellular reprogramming research.
They claimed to be able to reprogram mouse B lymphocytes into myeloid macrophage cells. They accomplished this by genetically forcing B lymphocytes to produce either C/EBPα or C/EBPβ proteins (5).
Later, they found that the exogenous introduction of C/EBPα into human, malignant B-lineage lymphoma cell lines, converts them into macrophage-like cells and prevents their further cell division. Artificial expression of C/EBPα basically turns off the tumor generating potential of these tumor cells! (6)
Inducing the differentiation of malignant cells is a common therapeutical strategy, e.g. in acute promyelocytic leukemia patients, e.g. by application of the vitamin A derivative all-trans-retinoic acid. Provoking transdifferentiation of tumor cells to silence their oncogenic potential, however, was definitely a completely novel approach to cancer therapy.
It happened to be that I needed to validate these amazing properties of C/EBPα or C/EBPβ in my own experimental work for one specific question which, however, I will not further address.
This is getting long now and I will make a cut here. Please stay tuned for part II where I will show you the data that I acquired to test these C/EBP proteins myself.
Yours, @replichara
Literature:
(1) https://en.wikipedia.org/wiki/C._H._Waddington
(2) Tapscott, S.J., Davis, R. L., Thayer, M.J., Cheng, P.F., Weintraub, H. & Lassar, A.B. (1987) MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science, Oct 21, Vol. 242(4877):405-11.
(3) Pabst, T., Mueller, B.U., Zhang, P., Radomska, H.S., Narravula, S.,Schnittger, S., ..., Tenen, D.G. (2001) Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nature Genetics, Mar 27, 27(3): 263-270.
(4) Familiades, J., Bousquet, M., Lafage-Pochitaloff, M., Béné, M.C., Beldjord, K., De Vos, J., ..., Delabesse, E. (2009) PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia, Nov 23, 23(11): 1989-1998.
(5) Xie, H., Ye, M., Feng, R. & Graf, T. (2004) Stepwise reprogramming of B cells into macrophages. Cell. May 28;117(5):663-76.
Image Sources:
-1- Pixabay
-2- Sandoval, S., Torres, C., García, M.P., Aldana, M. (2014). Criticality in gene networks. Frontiers in Ecology, Evolution and Complexity, 159-177.
-3- Mikael Häggström, from original by A. Rad. Creative Commons Attribution-Share Alike 3.0 Unported license.
-4- RCSB Protein Data Bank, entry: 1NWQ.
Fantastic post! My field is in immunology, so I read and write a lot about subjects that people like to discredit the science behind :)
I really look forward to seeing more of your series, as I believe that laying out the whole hellish process of scientific discovery is essential to differentiate between Google search research and....stuff that gets published. Resteemed!
Thanks a lot for the support. I read your recent article. It's on spot! Going to upvote it when my voting power is restored.
I am not sure if I can make a real series out of this, but maybe more fellow Steemit scientists could join in! Scientific discovery is a really painstaking process that many people, my earlier self included, greatly underestimate.
Thank you so much! I would be really interested in collaborating and contributing to this series...an anthology of scientific discovery perhaps?
Excellent post, I enjoy how you go through the evolution of this particular field of science. Looking forward to the next one!
Thank you! And welcome to Steemit. We need more professional academics in here :-)
Now, I'm already anticipating a good deal of scrutiny towards the second part of the article, haha!
"To truly understand something, you have to exercise and perform it practically - yourself" This on i will remember
This post has received a 0.14 % upvote from @drotto thanks to: @steemstem-bot.
Well done! This post has received a 7.14 % upvote from @litasio thanks to: @steemstem-bot. Whoop!
If you would like to delegate to the @LitasIO you can do so by clicking on the following link: 10SP
Congratulations @replichara, this post is the most rewarded post (based on pending payouts) in the last 12 hours written by a User account holder (accounts that hold between 0.1 and 1.0 Mega Vests). The total number of posts by User account holders during this period was 3664 and the total pending payments to posts in this category was $10074.69. To see the full list of highest paid posts across all accounts categories, click here.
If you do not wish to receive these messages in future, please reply stop to this comment.
Great post.
Thanks. Glad you liked it.
You are welcome