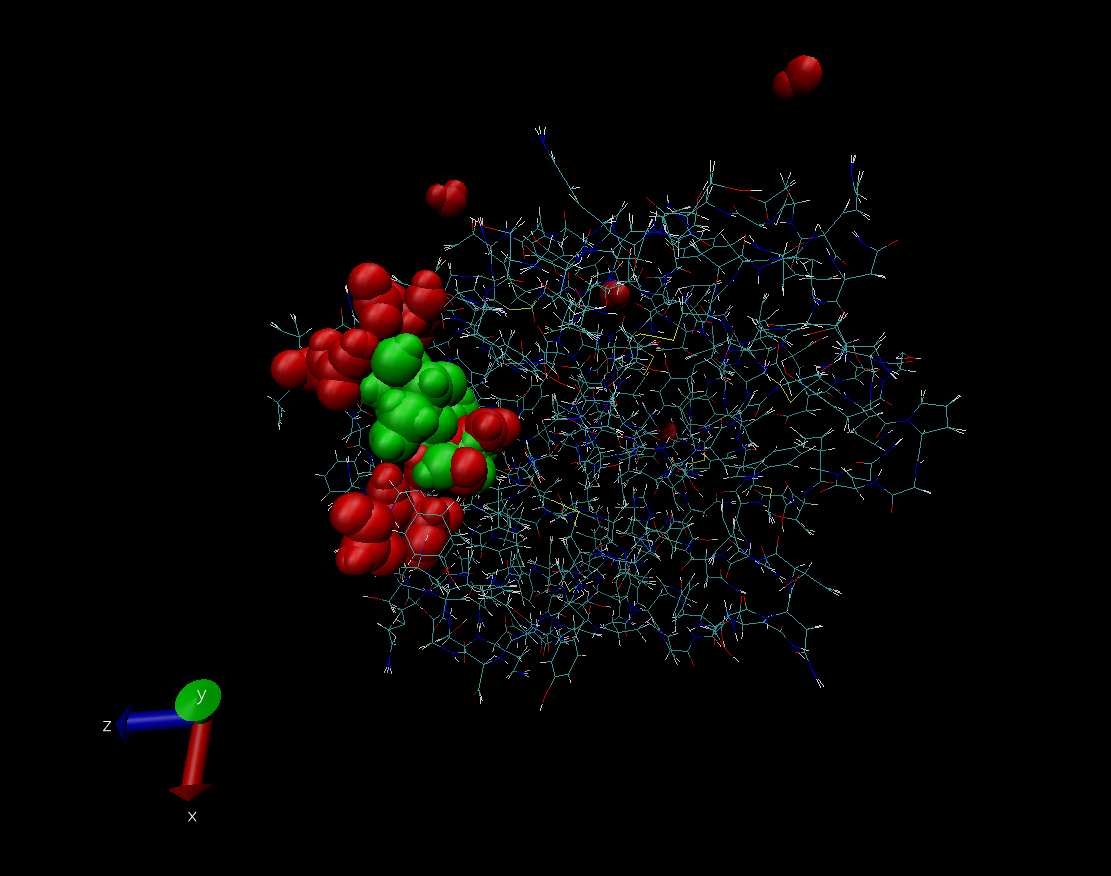
Molecular Dynamica: Docking a sucrose molecule onto a trypsin protein solvated in water
Over the past summer I helped out on a computational chemistry/biology model of sucrose bound to trypsin. Trypsin is the biological protein molecule of interest. The thermodynamic stability of the trypsin molecule was first tested by increasing the temperature incrementally over the course of a simulation with trypsin solvated in water. The denaturation of the protein was determined by taking quantitative snapshot analyses of the protein throughout the simulation. This data was then compared to a system with the trypsin molecule solvated in water, but with a sucrose molecule bound to a binding site (not active) on the outside of trypsin. In past chemical research the addition of sucrose to the system has increased the thermodynamic stability of the trypsin protein. The actual docking of sucrose to trypsin was a very time intensive task using the packmol program. The coordinates of all atoms in the binding site were used to match up to the specific atoms in sucrose that bind together. However, results of our computational model are still being analyzed at various temperatures. Results for this theoretical experiment will be posted soon. Here is a picture of sucrose bound to the the outside of a trypsin molecule.